

The development of science can be compared to picking mushrooms. For a long time, a mushroom picker can walk through the forest with very meager prey. But it is worth coming across a “mushroom place”, and the adept of quiet hunting is provided with full baskets of trophies that cause envy among friends and fellow travelers. Moreover, now the mushroom picker will return to the treasured clearing more than once, finding there more and more new crops.
In science, either new methods of interpreting data become such mushroom places, as the Pythagoreans achieved considerable success in mathematics, but without access to the positional number system, they were forced to be content with geometric and musical expressions of their theories, or new methods of obtaining data. They will be discussed further.
Each time, the appearance of a new method is akin to the appearance of a new sense organ in scientists. In biology, the latest case of such improvement of researchers at the moment was the advent of the sequencing method. Thanks to him, we now know what the actual structure of biomolecules is.
However, for modern biologists, this is no longer a luxury, but a matter of course. But the mushroom place continues to delight researchers with more and more new versions of this, without exaggeration, key technology for modern science. We will talk about three of them today. (We recommend the article: "Modern approach to the cultivation of microorganisms and cell cultures")
Transcriptome profiling
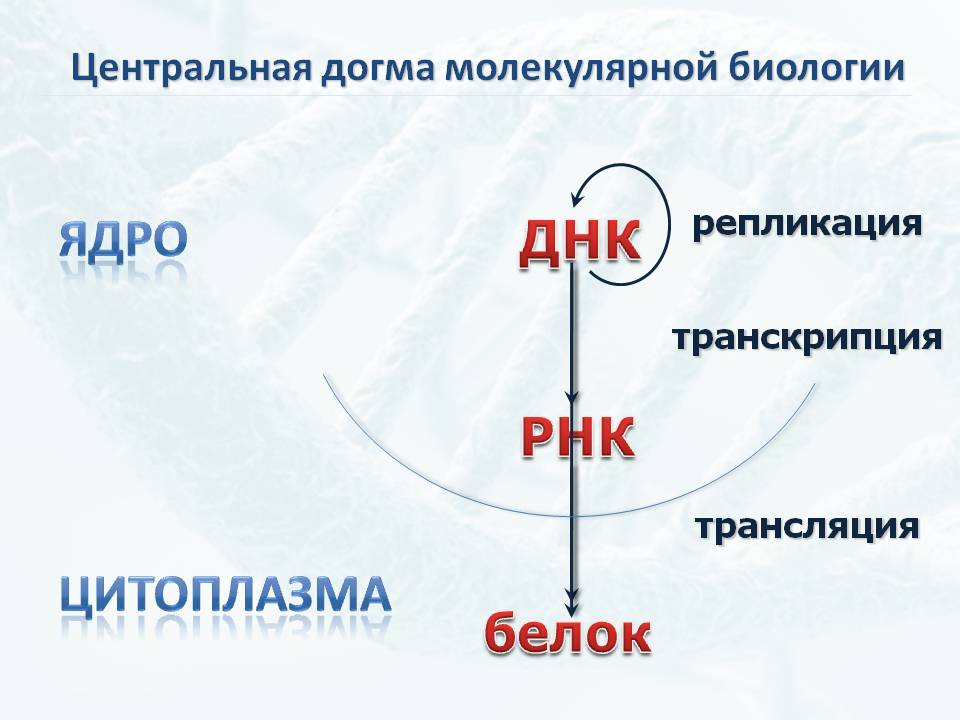
The basic postulate of molecular biology says that the information recorded in the genetic code of a cell is realized in the form of protein synthesis, through RNA. That is, first in the process of transcription, an RNA copying the gene appears. And already on RNA in the process of translation, protein is synthesized.
At the same time, proteins are extremely diverse, and their isolation from the cell can cause methodological difficulties. But RNA, even if there are three types of them, transport, information and ribosomal, is quite homogeneous in its properties. So the isolation of the RNA contained in the cell, although it has certain difficulties, has already become a well-established, routine process.
All RNA in a cell is called a transcriptome, and it can be used to judge what exactly the cell is doing. Moreover, this assessment is strictly quantitative. Transport or ribosomal RNA directly show how active the processes of protein synthesis are. At the same time, the definition of messenger RNAs gives a much more differentiated idea of which proteins are synthesized in this part of the body. Innovative equipment makes it possible to evaluate these processes even in single cells.
The process is more lengthy than complex. True, it is necessary to already have an annotated reference genome of the organism under study. Well, then it's a matter of technology. And in the truest sense of the word: most procedures are performed by special programs. The result of their work is the number of RNA reads,

corresponding to different regions of DNA. In simple terms, this shows which genes are active and involved in protein synthesis at the moment. Well, already by what these proteins are, what their function is, we can conclude what functions the cell as a whole performs.
In addition to scientific problems, this method also allows solving purely practical problems. Determining the functionality of a tissue or organ with transcriptome profiling becomes not just a matter of clinical observation, but a completely objective indicator. There is a wide range of equipment to solve these problems.
However, transcriptome profiling also has even more subtle variations that allow solving even more specific problems. For example, not just detecting actively transcribed genes, but also more productive alleles, or identifying alternative splicing. The latter became especially relevant in the study of squids, when it turned out that, unlike many other organisms, they do not have a one-to-one correspondence “one gene – one protein”. On the contrary, the use of different variants of splicing, i.e. excision of non-coding regions from RNA, allows a small number of intersecting regions of the genome to encode a significant number of proteins.
High throughput genotyping
Remember how we said above that successful transcriptome profiling requires an annotated reference genotype? So, obtaining it for a variety of organisms is now one of the most urgent tasks of molecular biology.
However, if the only way to determine the DNA sequence until now was the most common Sanger sequencing in laboratories, the genome of even the simplest organism would have to be deciphered for years. In fact, it took 13 years to read the human genome. And this despite the fact that over the course of the project, more and more advanced methods and equipment appeared, and hundreds of laboratories around the world were involved in this project. (We recommend the article: "Innovative technologies for the search and development of new drugs")
However, Sanger sequencing is not in vain remains the gold standard for determining the sequences of molecules that store genetic information. The reason for this is its extremely high accuracy and relatively uncomplicated automation. Well, speed can sometimes be sacrificed. Even in our fast-paced age, scientific research is not the fastest process.
However, the improvement of methods and equipment does not stand still. One of the latest developments in the field of reading nucleic acid sequences has been high throughput sequencing.
Without going into technical details, the essence of the method is that the genome is divided into many fragments, each of which enters its own microscopic reactor. This is possible by creating an emulsion with the smallest nanosized drops, each of which contains the substances necessary for the reaction. And so, in the beginning, each DNA fragment is accumulated due to the polymerase chain reaction, and then it is immobilized and goes through the process of almost ordinary sequencing.


The difference with the traditional method is that the attachment of each individual nucleotide is recorded. Recall that in the standard method, the entire genome region is first synthesized, but after each nucleotide, some of the reaction products no longer grow. Thus, the result is a mixture of DNA segments of all possible lengths. They are then separated by electrophoresis. Here, everything happens the other way around. Each time, only one kind of nucleotide is fed into the reaction mixture, and if it is suitable to attach to the growing copy of DNA, the accompanying reagents cause the glow.
That is, in this technique, the sequence of a large number of genome regions is determined at once, and sequence identification does not require an additional indication step.
Chromosomal microarray analysis
It is somewhat reminiscent of the previous technique and the now quite popular procedure of chromosomal microarray analysis. Although it cannot be attributed to sequencing. In addition to the sequence of the genome, an important characteristic of human health is the absence of violations in its chromosomes, doubling or disappearance of individual sections.
The essence of the method is quite simple. The basis of the technology is the use of microarrays. These are special chips with small sections of DNA fixed on them, corresponding to different areas of the human genetic code. For analysis, two types of genetic material are used - the patient himself and a standard template corresponding to a healthy person. Both materials are fragmented and marked with luminous markers.

During the analysis, fragments of the studied and standard DNA are applied to a microarray. Both of them bind to fixed complementary regions, and then they simply evaluate which of the signals, analyzed or standard DNA, is stronger in each region of the microarray. In the absence of anomalies, they will not differ. If in the genetic material there is a shortage, or vice versa, an excess, of any part of the chromosome, then in this section of the micromatrix the signal of the analyzed sample will be weaker, or, accordingly, stronger.
However, this is not the only application of this method. If you replace the standard DNA with some other, for example, a potential parent of the patient, then you can find not anomalies, but similarities and differences in the genome of two people - that is, to confirm the relationship.
And this concludes the consideration of new methods of genetic analysis. This is a very vast and interesting area of human knowledge, but it is impossible to delve deep enough into all the variety of methods and technical findings used by molecular biologists today in such a short article.

